In recent years, gene editing represented by CRISPR / Cas has brought revolutionary progress in biotechnology. In 2015 and 2017, Science was awarded as the first of the annual breakthrough scientific progress, the first of the five most influential scientific events in Nature in the past 10 years and the 2020 Nobel Prize in Chemistry. In just a few years, CRISPR technology has become a popular content in the scientific research community, known as the next generation of molecular diagnostic technology!
CRISPR / Cas system introduction
In 1987, the CRISPR locus was found in the bacterial genome, and CRISPR was found in 2002. It was subsequently confirmed that the CRISPR-Cas system is an RNA-guided adaptive immune system that can resist viruses and plasmids. The immune process can be roughly divided into three stages: adaptation, expression and interference.
(1) In the adaptation stage, Cas proteins recognize and capture foreign nucleic acid fragments, acquire new spacer sequences, and integrate and insert them into their own CRISPR array to form immunological memory.
(2) expression stage when foreign nucleic acid invasion again, corresponding from the CRISPR array, transcribed CRISPRRNA (crRNA) precursor and processing of small, mature crRNA, containing a conserved repeat sequence and a spacer sequence, crRNA further interaction with one or more Cas proteins, forming the RN (ribonucleoprotein) complex.
(3) The Cas-crRNA complex recognizes foreign nucleic acids and mediates the specific destruction of invading nucleic acids by targeting the target nucleic acid region by crRNA.
CRISPR / Cas can be divided into two groups: Class 1 is characterized by effector complexes of multiple Cas, including I, and type; Class 2 requires Cas only a single multidomain Cas, including (Cas 9), V (Cas 12) and (Cas 13). Class 2 system is simple, efficient and easy to operate, and is the main gene editing system, among which the most representative is Cas 9.
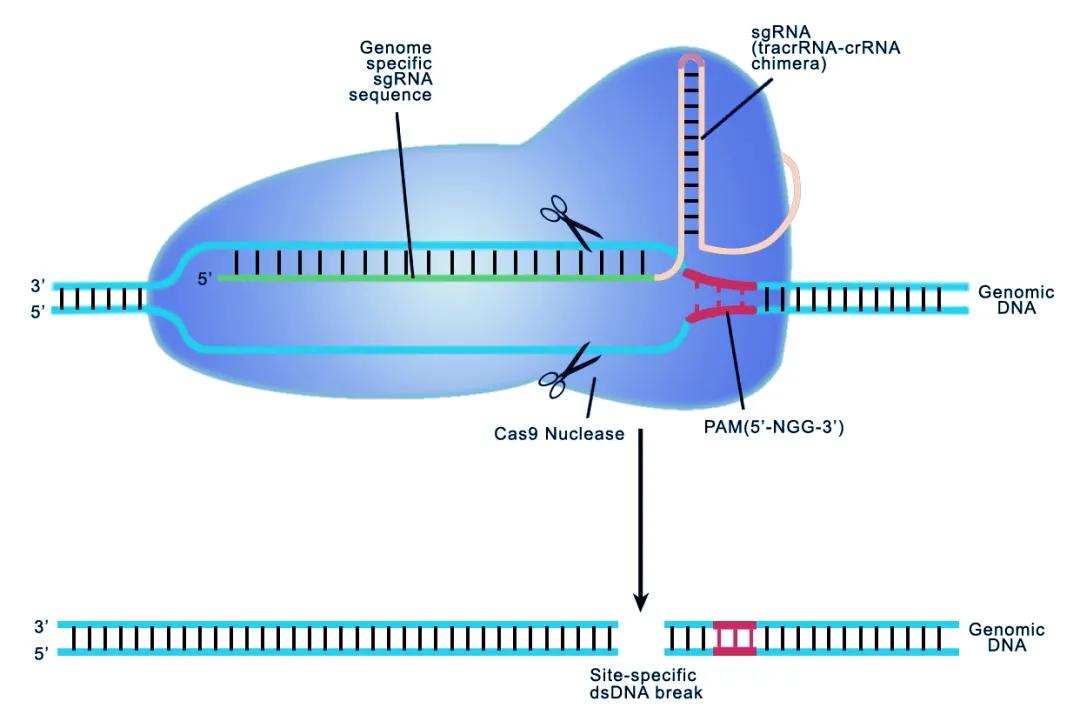
Cas 9 editing gene principle
Compared with Cas 9, Cas12a and Cas13a require specific target site cleavage, indicating that their system is more concise and has the potential to become a new generation of gene editing tools with higher accuracy and better safety.
A molecular detection system based on the Cas12a
Three Cas12a proteins widely used for nucleic acid detection include LbCas12a, AsCas12a, and FnCas12a, with LbCas12a having the strongest trans-cutting activity. In 2017, the Jennifer Doudna team developed the DETECTR (DNA endonuclease-targeted CRISPR trans reporter) detection technology, Using the LbCas12a (Lachnospiraceae bacterium ND2006), complexed with the crRNA, And use crRNA guidance to cut target DNA 18 to 25 nt downstream of the PAM (TTTN) sequence, While recognizing and cleaving the target dsDNA, To iting the trans-cutting activity of Cas12a, The ssDNA reporter molecule in the cleavage reaction system. The ssDNA reporter molecules are labeled with fluorescent and quenching groups, which generate a stable and intense fluorescence signal with an intensity an indicator of the amount of the target present in the detection sample. In addition, DETECTR is combined with the recombinant enzyme polymerase isothermal amplification technology (recombinase polymerase amplification, RPA), which not only improves the sensitivity of the assay (A mol/L level), but also avoids the need for complex and expensive equipment. This technique has now been successfully used in the detection of human papilloma virus (human papilloma virus, HPV) and SARS-CoV-2.
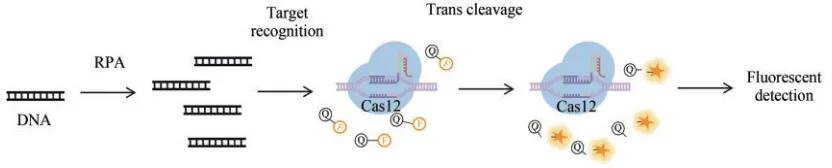
DETECTR A Schematic diagram of the detection method
In 2018, the team of Wang Jin and Zhao Guoping established the HOLMES (one-hour lowcost multipurpose highly efficient system) detection technology, Combined LbCas12a with PCR amplification, Can be done within 1 h (pseudorabies virus, PRV) and the Japanese encephalitis virus (Japanese encephalitis virus, JEV) detection, Sensitivity can be achieved up to 1 to 10 a mol/L, It also accurately distinguishes between viral genotypes and human single-nucleotide polymorphisms. However, this technique combined with PCR amplification to improve the stability but also increase the detection time and device dependence.
A molecular detection system based on the Cas12b
The Cas12b derived from hyperthermophiles contains a single RuvC domain, with a low off-target effect and also has trans-cleavage activity. CRISPRCas12b Is a dual RNA-guided DNA endonuclease system that can recognize and cleave targeted dsDNA or ssDNA or also nonspecifically cleave ssDNA. When targeting dsDNA, Cas12b relies on the PAM sequence (TTC or TAC) to complete cis-cleavage. When targeting ssDNA, Cas12b achieves cleavage independent of the PAM sequence and has higher cleavage activity than dsDNA.
According to these characteristics, The team of Wang Jin and Zhao Guoping established the upgraded HOLMESv2, Isothermal amplification of AacCas12b (Alicyclobacillus acidoterrestris) protein with loop-mediated (loopmediated isothermal amplification, LAMP) combine, Only a dsDNA containing 5 ʹ -TTC-3 ʹ or a DNA sequence with 5 ʹ -TAC-3 ʹ after cleavage could activate the AacCas12b trans-cleavage activity, This technique enables DNA and RNA-specific detection as well as discrimination between SNP and quantitative DNA methylation.
In 2019, Dai et al. developed the E-CRISPR detection technology based on CRISPR-Cas12a to influence the electrical signal output through the CRISPR-Cas system, and obtained the high conduction signal by using the trans-cutting activity of Cas12a. The detection technology fixed the ssDNA reporter molecule modified with a 3 ′ -methylene blue (3 ′ -MB) tag on the gold electrode. In the presence of the target DNA, Cas12a-crRNA cuts the MB-ssDNA molecule in trans, and the separation of the methylene blue tag will reduce the electrochemical signal. The trans-cleavage activity of Cas12a remains silent in the presence of no target DNA and the ssDNA molecules remain on the electrode surface, resulting in a high electrochemical current in methylene blue.

SHERLOCK Principle of the detection method
In 2020, Ding et al. developed AIOD CRISPR (all-in-one dual CRISPR-Cas12a) detection technology, which mixed all reaction components of RPA amplification and CRISPR detection and added high concentration of ssDNA-reporter molecules while introducing two crRNA, thus testing SARS-CoV-2 and HIV-1 in one step.
Molecular detection system based on Cas 13
Cas 13 is an RNA guide and RNA-targeted nuclease with two HEPN domains that act specifically on RNA targets. In 2017, Zhang feng's team developed the SHERLOCK (specific high-sensitivity enzymatic reporter unlocking) nucleic acid detection technology. This technique has the same principle as DETECTR, but relies on the activity of LwaCas13a (Leptotirichia wadei) nuclease to recognize and cleave target RNA while they cleave ssDNA reporter molecules in trans.
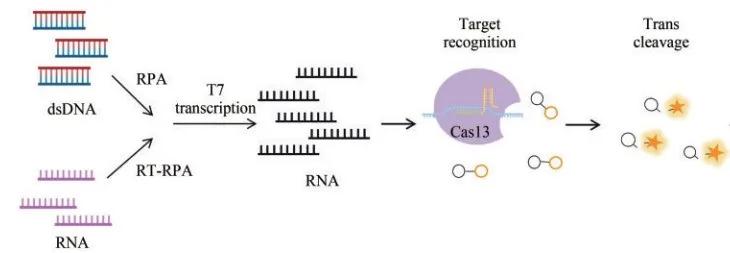
SHERLOCK Principle of the detection method
To address the quantification issue, the team developed SHERLOCKv2, which used a combination of four types of Cas proteins (LwaCas13a, PsmCas13b, CcaCas13b and AsCas12a), cut reporter molecules labeled with different fluorescent groups and tested in FAM, Cy 5, HEX and TEX channels, enabling quantification and simultaneous detection of four nucleic acid sequences. SHERLOCKv2 Was also applied to lateral flow analysis to detect the cleaved reporter molecule on a strip strip by gold nanoparticle-labeled anti-FAM antibody, generating a visual colorimetric signal.
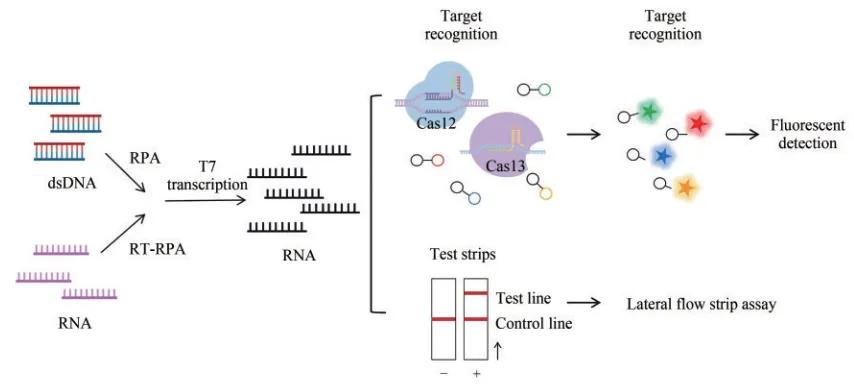
SHERLOCKv2 Reading method of the test results
In 2020, ACKERMAN and others combined CRISPR diagnostic and microfluidic technologies to develop the CARMEN (combinatorial arrayed reactions for multiplexed evaluation of nucleic acids) detection platform, which can detect eight samples and 169 human-related viruses simultaneously. However, due to the high cost of chip customization and the replication of the inspection operation process, it is difficult to apply in clinical practice. The team is at CARMENv.1 Based on the development of mCARMEN (microfluidic combinatorial arrayed reactions for multiplexed evaluation of nucleic acids), which combines CRISPR diagnosis and microarray technology with a simplified clinical workflow, the mCARMEN respiratory virus detection panel can simultaneously detect 21 respiratory viruses, including SARS-CoV-2 and other coronaviruses and influenza viruses.
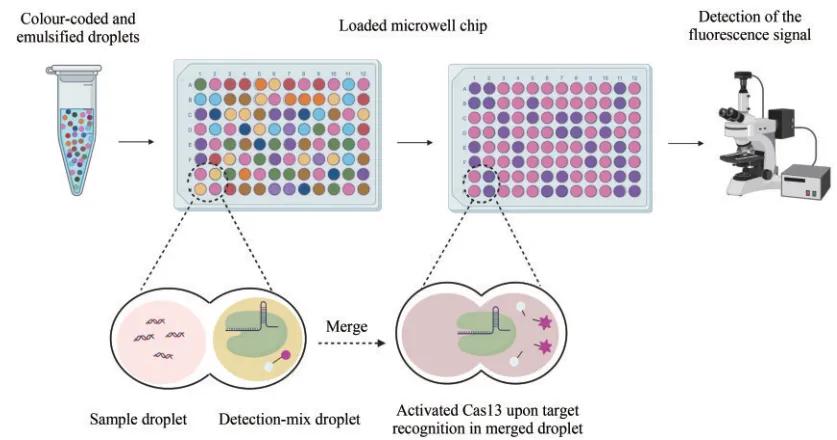
Principle of the CARMEN-Cas 13 assay
The Arizti-Sanz team integrated amplification with CRISPR-Cas 13 system detection into a one-step diagnostic platform SHIN (streamlined highlight of infection to navigate pandemic), which improved the sensitivity of nucleic acid detection by adding RNase H, and adopted in-tube fluorescence and matching smartphone application to achieve effective nucleic acid detection of SARS-CoV-2. The SHINE diagnostic platform minimizes device requirements and also reduces outcome reading bias.
Molecular detection system based on Cas 14
In addition to the Cas 12 and Cas 13 systems, the compact Cas 14 proteins are known as having similar trans-cleavage activity. Cas 14, a CRISPR endonuclease targeting ssDNA, Cas 14 recognizes and cleaves target nucleic acid sequences unconstrained by the PAM sequence and is less tolerant to nucleotide mismatches between crRNA and target templates and the internal sequence is more sensitive to nucleotide mismatch, a feature that enables Cas 14 to achieve high-fidelity discrimination SNP. The Cas14a-DETECTR detection platform established based on this feature has been applied to the SNP typing of the human HERC 2 gene.
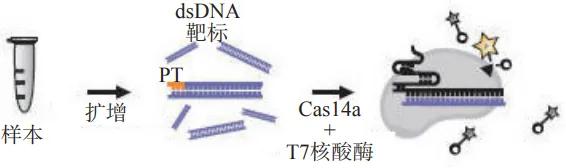
A Schematic diagram of the molecular detection of CRISPR / Cas14a
CRISPR / Cas technical advantages
So present, CRISPR-based nucleic acid detection technology has the following advantages:
(1) High specificity, enabling single-nucleotide point mutation detection;
(2) High sensitivity, which can realize single-copy nucleic acid detection;
(3) Rapidity, the whole test is completed within 0.5–1.0h;
(4) Simple, do not need professional equipment and personnel, can be completed on site;
(5) Reagent is resistant to freeze-drying and easy for storage and carrying;
(6) Easy to develop, can quickly develop new nucleic acid detection kits, etc.
With further improvement and optimization, it is more universal, and the CRISPR nucleic acid detection technology will be the most preferred technology for POCT.
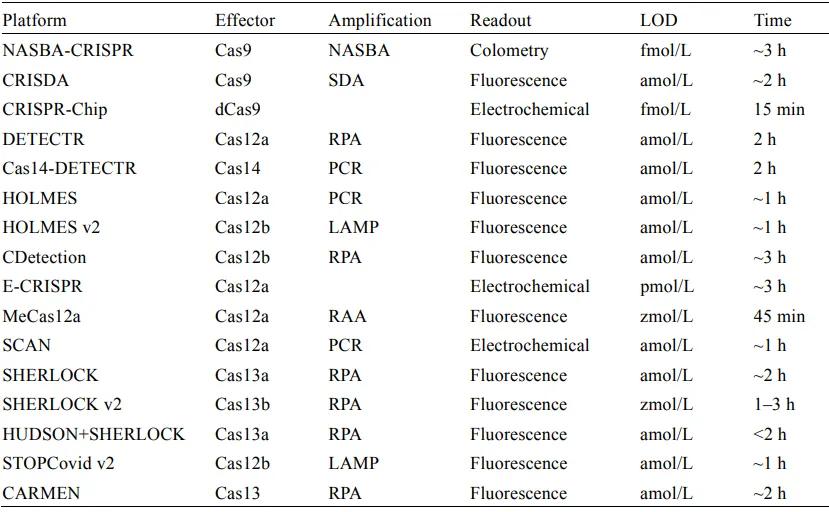
Comparison of the multiple CRISPR-based molecular detection techniques that have been reported
CRISPR / Cas future direction
First, the development of new Cas proteins and the screening and identification of mutants. For example, the later discovered Cas 14 protein, different from Cas 12 and Cas 13, is good at recognizing single-stranded DNA and has enriched the CRISPR toolbox. The screening and identification of Cas proteins with different cleavage characteristics provides a method to improve the detection flux and reduce the "off-target effects".
Second, it is to combine CRISPR / Cas and other technology platforms to develop novel and practical biosensing strategies. Such as the fixation of the Cas 9 protein to a graphene field-effect transistor and the reporter probe of the Cas system to the electrodes to build electrochemical biosensors that do not require signal amplification. Use the hydrogel as its response element to achieve a diversified signal output. The CARMAN technology developed in combination with microfluidic technology can quickly detect dozens of samples and hundreds of viruses, providing a method to solve the problem of low throughput.
reference documentation
[1] Du Yao, Gao Hongdan, Liu Jiakun, Liu Xiaorong, Xing Zhihao, Zhang Tao, Ma Dongli. Progress of CRISPR-Cas system in pathogenic nucleic acid detection [J]. Synthetic biology, 2024,5 (1): 202-216.
[2] Sun Wenjun, Huang Xingxu, Wang Xinjie, et al. Rapid, sensitive and convenient molecular detection based on CRISPR. Journal of Bioengineering, 2023,39 (1): 60-73.
[3] Wang Xueliang, Xiao Yanqun, Wang Jialiang. Application of the CRISPR / Cas system in molecular detection [J]. Laboratory Medicine, 2020,35 (2): 181-185.
[4] MAKAROVA K S, ZHANG F, KOONIN E V.SnapShot: class 1 CRISPR-cas systems[J].Cell, 2017, 168(5): 946-946.e1.
[5] SHMAKOV S, ABUDAYYEH O O, MAKAROVA K S, et al.Discovery and functional characterization of diverse class 2 CRISPR-Cas systems[J].Molecular Cell, 2015, 60(3): 385-397.
[6] JINEK M,CHYLINSKI K,FONFARA I,et al.A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity[J].Science,2012,337 (6096):816-821.
[7] LEUNG R K K, CHENG Q X, WU Z L, et al.CRISPR-Cas12based nucleic acids detection systems[J].Methods, 2022, 203: 276-281.
[8] LI S Y, CHENG Q X, WANG J M, et al.CRISPR-Cas12aassisted nucleic acid detection[J].Cell Discovery, 2018, 4: 20
[9] ENG F, CUI T T, FENG G H, et al.Repurposing CRISPRCas12b for mammalian genome engineering[J].Cell Discovery, 2018, 4: 63.
[10] TENG F, GUO L, CUI T T, et al.CDetection: CRISPRCas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity[J].Genome Biology, 2019, 20(1): 132.
[11] KELLNER M J, KOOB J G, GOOTENBERG J S, et al.SHERLOCK: nucleic acid detection with CRISPR nucleases [J].Nature Protocols, 2019, 14(10): 2986-3012.
Our company's product recommendation:
1.5678-48-8 https://www.bicbiotech.com/product_detail.php?id=5405
2.5678-45-5 https://www.bicbiotech.com/product_detail.php?id=5406
3.6049-54-3 https://www.bicbiotech.com/product_detail.php?id=5407
4.68208-19-5 https://www.bicbiotech.com/product_detail.php?id=5408
5.325-89-3 https://www.bicbiotech.com/product_detail.php?id=5409




