background
Gene therapy is a means of introducing genetic material that changes the function of specific cells into patients to treat genetic diseases. A critical step in gene therapy is the efficient delivery of genes to target tissues / cells by gene delivery vectors. Currently, gene passage vectors are divided into two categories: viral vectors and non-viral vectors. Currently, viral vector-based gene therapy is mainly achieved through the delivery of therapeutic genes to patients by retrovirus, adenovirus (Ad), or adeno-associated virus (AAV) vectors (Figure 1). The lack of adequate understanding of viral biology has led to poor virus-based gene therapy.

Figure 1. Diagram-representation of the viral gene therapy approach.
In February, 2021, the Jote T. In a review called "Viral vector platforms within the gene therapy landscape" in Signal Transduction and Targeted Therapy journals, Bulcha1 et al, In that review, The authors detailed three viral vector platforms, Most gene therapy vectors are based on Ads, AAVs, and lentiviruses (retroviruses), This article will describe the structure and route of infection of the above three viruses, To help you better understand the role of viral vectors in gene therapy.

contents of article
Generally, a viral vector is composed of three components: (1) the protein capsid and / or envelope wrapping the genetic payload and defining the tissue or cell affinity and antigen recognition of the vector; (2) the target gene that can have the desired effect when expressed in the cell; and (3) the regulatory cassette, a combination of enhancer / promoter / accessory elements that controls the expression of the transgene as a stable or transient cell of free or chromosomal integrons. The design of these three components of each viral vector platform is different, with their own strengths and weaknesses.
1 Adenovirus vector (Adenovirus vectors) |
Large-volume, transient target gene delivery
The Structure and the Genomes:
Ads are a nonenveloped virus that primarily causes upper respiratory tract infections and can also infect other organs such as the brain and bladder. It has an icosahedral protein capsid that houses anywhere from 26 to 45 kb of a linear double-stranded DNA genome. The Ads genome is flanked by hairpin-inverted terminal repeats (ITRs) of different lengths (terminal 30,371 bp). ITRs as self-initiating constructs can promote primase-independent DNA replication. Viral genome packaging requires a packaging signal located on the left arm of the genome. The Ads genome encodes about 35 proteins that are expressed early and late stages of viral gene transcription. The Ads genome includes five so-called "early" gene compositions, namely E1A, E1B, E2, E3, and E4 (Figure 2).
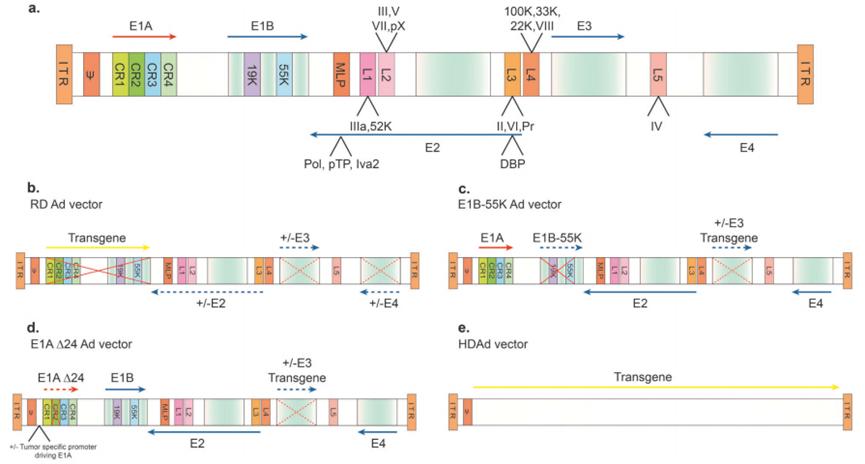
Figure 2. Schematic representation of the genome of wild-type adenovirus type 5 (Ad 5) and common Ad 5-based vectors.
route of infection:
To date, more than 100 human Ad genotypes have been identified, divided into seven subgroups from A to G, and the knowledge of the Ad infection pathway is mainly based on human Ad 5 (HAd 5). Infection begins with an interaction between the cell surface-localized coxsackievirus-Ad receptor (CAR) and the distal domain of the viral capsid fiber. In addition, many other Ad receptors have been identified, such as CD46, DSG 2, and sialic acid. Virus binding to cell surface receptors and endocytosis is mediated by the interaction between the tripeptide Arg-Gly-Asp (RGD) motif on the penton and α V integrins on the host cell surface. After endocytosis, the capsid is disassembled and the V and VI proteins promote endosomal escape. Viral DNA then enters the nucleus through the nuclear membrane pore, and in addition, the hexon of partially disrupted virions binds to dynein motors, helping the virus to transport to the nuclear pore through the microtubule network to enter the nucleus, and viral DNA remains mainly in the epichromosomal state and does not integrate into the host cell genome.
2 Adeno-associated viral vectors (Adeno-associated Virus vectors) |
A viral vector with immense potential
The Structure and the Genomes:
AAV was discovered in 1965 by Bob-Atchison (Bob Atchison) as a contaminant of an adenovirus agent. It is classified as dependent parvovirus because it relies on adenovirus or any virus that can exert an auxiliary function to complete its life cycle. AAV lacks the essential genes required for replication and expression of its own genome, and these essential functions can be provided by the E1, E2a, E4, and VA genes of Ad. The AAV genome itself is single-stranded DNA and contains four known open reading frames (ORFs).
The first ORF encodes four duplication genes (Rep), named after its molecular weight: Rep 40, Rep 52, Rep 68, and Rep 78. The second ORF encodes the three viral capsid proteins, VP1, VP2, and VP3, respectively. The third and fourth are nested subgenomic mRNA, named Assembly Activator protein (AAP), which is involved in the nucleolar shuttling of capsid monomers leading to capsid assembly; and the recently discovered membrane-associated accessory protein (MAAP) whose function is not fully understood. The genome is 4.7 kb long and is flanked by an ITR of 145 bases (Figure 3). ITRs serve as self-initiating structures for replication and provide signals for Rep-mediated packaging. The AAV capsid is a capsid of T=1, icosahedral, 60 polymer, with VP1, VP2 and VP3 subunits in a ratio of 1 ∶ 1 ∶ 10, respectively. The assembled capsid is best characterized by the formation of a facial five-fold symmetry axis of Rep binding and contains five pores, in which DNA is inserted into the capsid under Rep, mediated ATP enzyme activity; and a three-fold symmetry axis defined by triple protrusion including variable loop regions V, VII, VIII, and IV.
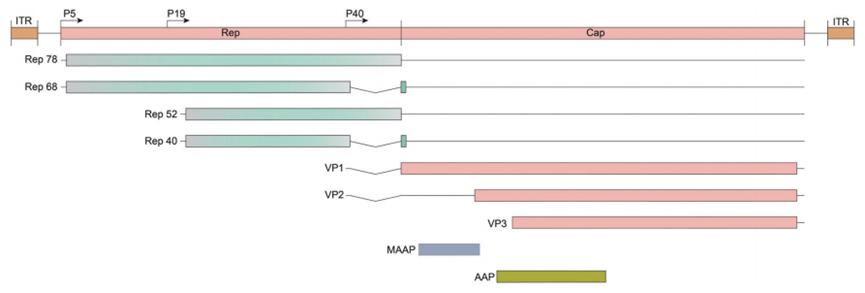
Figure 3. Schematic diagram of the AAV genome and the PCR screening loci.
route of infection:
Our knowledge of the route of AAV infection is based on only a few serotypes. The general understanding is that serotypes binding to surface glycoprotein receptors and secondary receptors of different cells may have different affinities. Upon binding to the cell surface, clathrin-mediated endocytosis is triggered and AAV particles are subsequently delivered into the endosomal vesicles and through the late endosomal and lysosomal region compartments. Due to the low pH environment of these vesicles, the capsid undergo conformational changes that expose the VP1 / 2 structure, the VP 1 unique region (containing the phospholipase domain) escapes the endosome or lysosomal compartment and then shuttles into the nucleus through nuclear localization signals found on VP2. Once in the nucleus, the AAV genome will synthesize the second strand, forming the double-stranded genomic structure required for gene transcription.
3 Lentiviral vector (Lentivirus vectors)|
A powerful vector for genetically modified cell therapy
The Structure and the Genomes:
Lentiviruses belong to retroviruses, which are spherical, enveloped, single-stranded RNA viruses with about 100 nm in diameter. Lentiviral particles wrap around two justice-chain RNA bound to nucleocapsid proteins. The particle also contains reverse transcriptase, integrase, and protease proteins. The lentiviral HIV-1, at 9.7 kb flanked by 5 ′ and 3 ′ LTR, dominates the viral genome replication. The LTR consists of unique U5 and U3 sequences, as well as R sequences shared by both ends. The cis-acting sequence, also known as psi, is located outside of the LTR. Retroviruses share common basic core protein genes, such as gag, pol, and env. The gag genes encode protective capsid and matrix proteins, the env genes carry information about the transmembrane and envelope glycoproteins that determine viral cell tropism, and the pol gene transcription reverse transcriptase and integrase. Lentiviruses have additional genes tat and rev, tat assists activation of transcription and RNA by binding to LTR, elongation of polymerase II. The rev coordinates the nuclear export of spliced and unspliced viral RNA by binding to motifs in environmental gene regions. Additional accessory genes, such as: vif, vpr, vpu, and nef, can increase viral titers and promote viral pathogenesis (Figure 4).
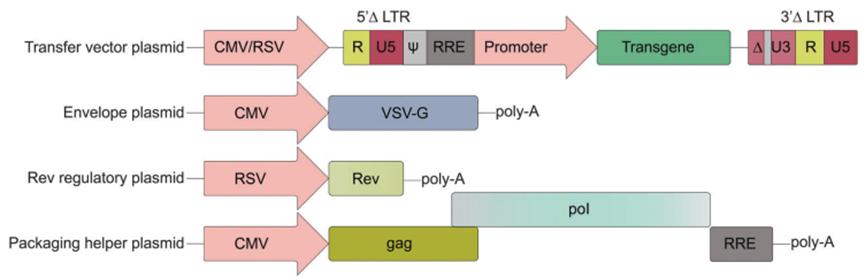
Figure 4. Third-generation HIV-1 lentiviral vector design.
route of infection:
The entry of lentiviral particles into host cells is mediated by interactions between glycoproteins anchored to the outer membrane and specific cellular receptors. Successful binding to cell surface receptors triggers a cascade of events that lead to the fusion of the viral particle lipid bilayer with the cell and the subsequent delivery of the viral genetic cargo to the cytoplasm. Lentiviral DNA is integrated into the host genome in a non-random manner and preferentially selects transcriptionally active sites.
discuss
In this review, the authors discuss the present situation of viral vectors, especially those based on Ad, AAV and lentivirus, points out that the future based on viral vectors is bright, the ability to solve many human genetic diseases can be realized, but the current method still has some shortcomings, need to further explore virus biology and interdisciplinary methods to overcome the current challenges.
reference documentation
Bulcha J T, Wang Y, Ma H, et al.Viral vector platforms within the gene therapy landscape[J].Signal transduction and targeted therapy, 2021, 6(1): 53.




