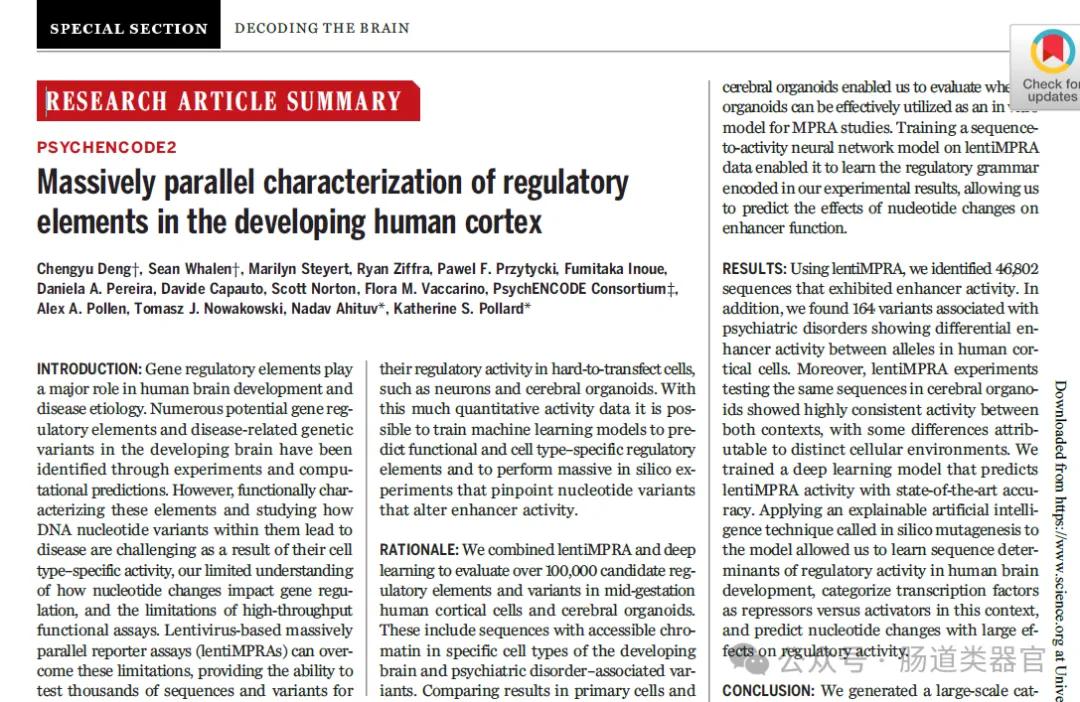
Gene regulatory elements play important roles in human brain development and disease etiology. Over the past few decades, scientists have identified through experimental and computational prediction methods many potential genetic regulatory elements and genetic variants associated with diseases in the developing brain. However, given their cell-type-specific activities, our limited understanding of how nucleotide changes affect gene regulation, combined with limitations of high-throughput functional detection, functional characterization of these elements and studying how DNA nucleotide variants cause disease becomes a major challenge.
In recent years, the development of massively parallel report analytics (lentiMPRA) has provided the possibility to overcome these limitations. This technology is able to test the regulatory activity of thousands of sequences and variants in difficult to transfect cells, for example, neurons and cerebral organoids. With large amounts of quantitative activity data, machine learning models can be trained to predict functional and cell-type-specific regulatory elements and to perform large-scale computer simulation experiments to identify nucleotide variants that alter enhancer activity.
Despite advances in the identification and functional studies of gene regulatory elements, remains to link link elements to specific cell types and their role in brain development. Moreover, our understanding of how genetic variation in noncoding regions affects gene expression and brain development through long-range regulatory mechanisms.
This study evaluated over 100,000 candidate regulatory elements and variants in human cortical cells and cerebral organoids in mid-gestation by combining lentiMPRA technology and deep learning. This includes sequences in accessible chromatin of specific cell types in the developing brain, as well as variants associated with psychiatric disorders. By comparing the results in primary cells and cerebral organoids, the investigators are able to assess whether the organoids can be effectively used as an in vitro model in MPRA studies. Furthermore, by training a sequential activity-based neural network model, the researcher is able to learn the regulatory grammar encoded in the experimental results to predict the effect of nucleotide changes on enhancer function.
This study not only provides new insights into understanding the regulatory code in human brain development, but also generates tools that can predict how nucleotide changes perturb regulatory elements. This has important implications for revealing how gene regulatory variants lead to phenotypic effects, while also providing potential targets for early diagnosis and treatment of psychiatric disorders.
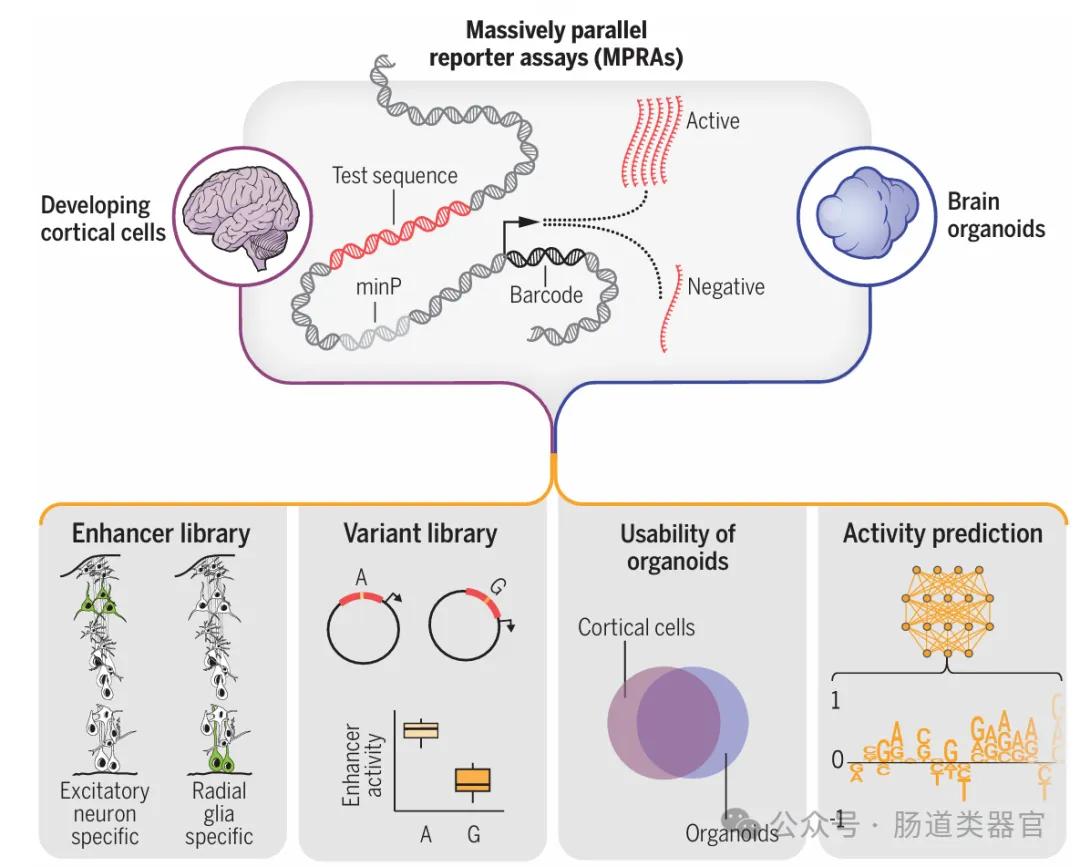
Gene regulatory elements play important roles in human brain development and disease etiology. Although existing studies have identified many potential gene regulatory elements and genetic variants associated with diseases in the developing brain through experimental and computational prediction approaches, the functional characterization of these elements and how DNA nucleotide variants cause disease remains a challenge. The study by Deng et al. performed a large-scale evaluation of regulatory elements and variants in second-trimester human cortical cells and cerebral organoids by combining lentiviral massively parallel reporter analysis (lentiMPRA) techniques and deep learning.
The team first developed the hypothesis that lentiMPRA technology can test the regulatory activity of a large number of sequences and variants in primary cells and brain organoids, and can use deep-learning models to predict functional and cell-type-specific regulatory elements. They designed two lentiMPRA libraries, one to characterize the regulatory potential of candidate cell type-specific enhancers and the other to compare the regulatory activities of reference and alternative alleles for 17,069 variants. Experimental results identified 46,802 active enhancer sequences in primary cells and identified 164 psychiatric-associated variants exhibiting allelic differences in enhancer activity in human cortical cells.
The key finding in the study is that cerebral organoids and primary cells exhibit highly consistent activity in lentiMPRA experiments, suggesting that organoids can serve as an appropriate model to studying the regulatory landscape of the developmental cortex. Moreover, deep learning models to predict lentiMPRA activity with state-of-the-art accuracy allow researchers to learn sequence determinants of regulatory activity in human brain development, classify transcription factors as repressors and activators, and predict nucleotide changes that have a significant impact on regulatory activity.
The results highlight the potential of brain organoids as research tools and reveal variants of gene regulatory activity associated with psychiatric disorders. The work of Deng et al not only advances our understanding of the regulatory code of human brain development but also generates tools that can predict how nucleotide changes perturb regulatory elements. These findings have important implications for understanding how gene regulatory variants cause phenotypic effects and provide potential targets for early diagnosis and treatment of psychiatric disorders.
However, the study also has several limitations. For example, MPRAs, despite measuring the regulatory activity of the sequence, do not identify their target genes. Furthermore, current methods have difficulty in detecting cell type-specific regulatory elements in mixed cell populations due to technical limitations. Nonetheless, this study provides new directions for future studies, including the use of single-cell technologies or purified cell populations to validate larger numbers of sequences, and using CRISPR screens to assess the endogenous activity of candidate regulatory sequences. The integration of these strategies will be a key step in deciphering the developmental regulatory code of the human brain.
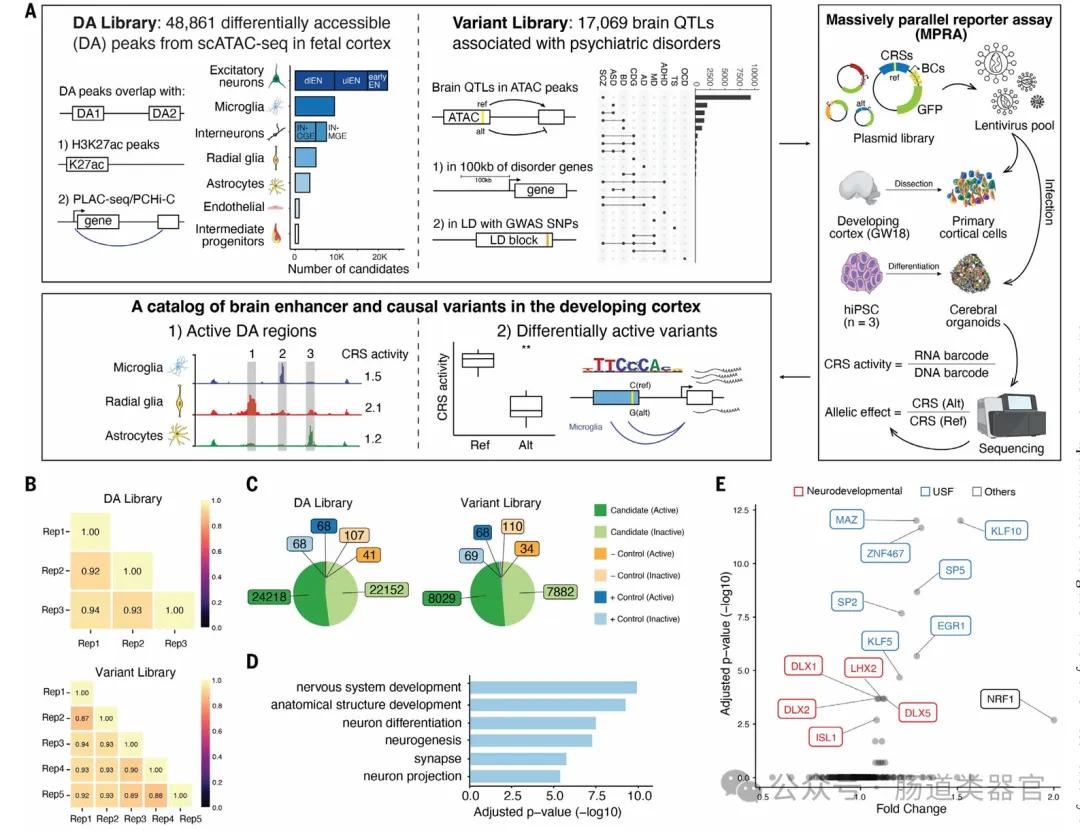
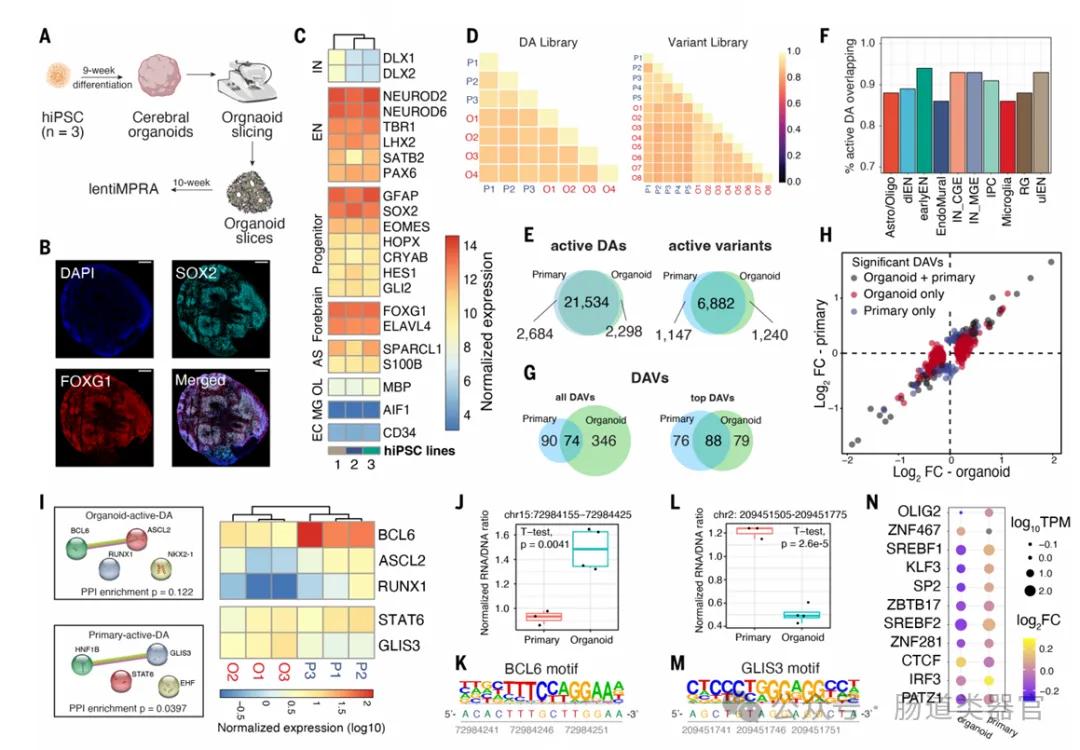
In this study, the application of cerebral organoids is critical as they provide an in vitro model to model the activity of the regulatory elements during human brain development. Organoids are three-dimensional cell cultures derived from pluripotent stem cells that can mimic the structure and function of the brain to some extent. The investigators used these organoids to test and compare the activity of gene regulatory elements in specific cell types in the developing brain.
Ten weeks old cerebral organoids were used in the study and their expression of expressed relevant cell type markers was verified by immunostaining and RNA sequencing. These organoids were cut into sheets and infected with lentiviral lentiMPRA libraries containing candidate regulatory sequences. In this way, the investigators were able to assess the enhancer activity of these sequences in organoids and compare them with the activity in primary cortical cells.
The results showed that organoids and primary cells showed a high degree of activity coherence in lentiMPRA experiments, suggesting that brain organoids can be used as an efficient in vitro model to study gene regulation in human brain development. Furthermore, the application of organoids has revealed several cell-type-specific regulatory elements that may be difficult to detect in primary cells because their activity in mixed cell populations may be diluted.
Furthermore, brain organoids are used to identify gene regulatory variants associated with psychiatric disorders. Researchers have identified several psychiatric disorder-associated variants in organoids that show interallele differences in enhancer activity, providing new insight into understanding how these variants affect brain development and the risk of psychiatric disorders.
Overall, the application of brain organoids in this study demonstrates their potential to model human brain development, identifying and characterizing gene regulatory elements, and studying genetic variation associated with psychiatric disorders. However, the investigators also note that although organoids are a useful model, differences in cell type composition and the context of transcriptional regulation need to be carefully considered when interpreting the lentiMPRA results. Future work may consider using organoid lentiMPRA methods to test libraries from different psychiatric disorders or nonhuman primates iPSCs, as well as using techniques such as single-cell MPRAs and CRISPR screening to further extend this study.

The innovation of this study is the combination of lentiviral massively parallel reporter analysis (lentiMPRA) technology and deep learning to explore and understand gene regulatory elements in the developing human brain. Here are a few innovations in the study:
1. The combination of high-throughput functional genomics and machine learning: Researchers have not only used lentiMPRA to evaluate more than 100,000 candidate regulatory elements and variants, but also developed deep learning models to predict regulatory activity, a combination that is not common in previous studies.
2. Prediction of cell type-specific activity: Through trained deep learning models, researchers can predict the regulatory element activity of specific cell types, which is important for understanding the regulation of specific gene expression in different cell types in the brain.
3. Large-scale catalog creation of gene regulatory elements: Research has generated a large-scale catalog of gene regulatory elements active in mid-gestation human cortical cells and cerebral organoids, which provides a valuable resource for future research.
4. Functional characterization of psychiatric disorders-related genetic variants: Studies identifying 164 variants associated with psychiatric disorders and analyzing their enhancer activity differences in human cortical cells can help reveal how these variants affect brain development and disease risk.
5. Validation of cerebral organoids as model systems: The application of cerebral organoids in research not only demonstrates their potential as a tool to study human brain development, but also verifies their effectiveness in simulating regulatory activity in primary cells.
6. Application of explanatory AI technology: By applying an explanatory AI technology called in silico mutagenesis, researchers are able to learn the sequence determinants of regulatory activity and predict nucleotide changes that have a significant impact on regulatory activity.
7. Prediction and classification of transcription factor functions: The application of deep learning models in research also allows researchers to classify the functions of transcription factors as activators or repressors in the context of a specific cell type.
The application of these innovative methods and techniques greatly advances our understanding of the function of regulatory elements in the developing human brain, providing new tools and strategies for studying the genetic basis of psychiatric disorders.
Although Deng et al. have made remarkable progress in the field of gene regulatory elements, there are still some shortcomings:
1. Cell-type-specific challenges: While lentiMPRA was used to assess cell-type-specific regulatory elements, the technology may have limitations in detecting rare cell types in mixed cell populations. This may result to some cell type-specific regulatory elements failing to be accurately identified.
2. Uncertainty of target genes: MPRAs technology can measure the regulatory activity of sequences, but usually do not directly identify their target genes. This limits a comprehensive understanding of the functional effects of regulatory elements.
3. Technical limitations: Although lentiMPRA technology is able to test multiple sequences with high throughput, it tests short sequences (270-bp), which may not be enough to capture regulatory elements that require a broader chromatin environment to achieve cell type specificity.
4. Differences in the context of transcriptional regulation: Although brain organoids have the potential to simulate human brain development, they differ from primary cells in cell type composition and the context of transcriptional regulation. This may influence the interpretation of the regulatory element activity.
5. Comparison with adult brain QTLs: QTLs in the adult brain, rather than QTLs in the developing brain, which may limit the discovery of genes related to neurodevelopmental disorders.
6. Correlation of effect size: The single nucleotide polymorphisms (SNPs) observed in studies usually have less effects on regulatory activity, which may limit their relevance in functional studies.
7. Experimental association with endogenous gene expression: MPRAs usually measure the activity of reporter genes, rather than the expression of endogenous genes, which may pose challenges when linking experimental results to endogenous gene regulation.
8. Complexity of data interpretation: Although deep learning models can predict the activity of regulatory elements, the interpretability of the models and the biological significance of the prediction need further experimental verification.
9. Experimental replicability and universality: The methods and findings in research need to be replicated in other laboratories and in different populations to ensure the universality and reliability of the results.
10. Ethical and safety issues: The use of human stem cell and organoid technologies involves ethical and safety issues that need to be carefully considered in research.
These shortcomings suggest problems to be addressed in future research and provide direction for improving existing techniques and methods.
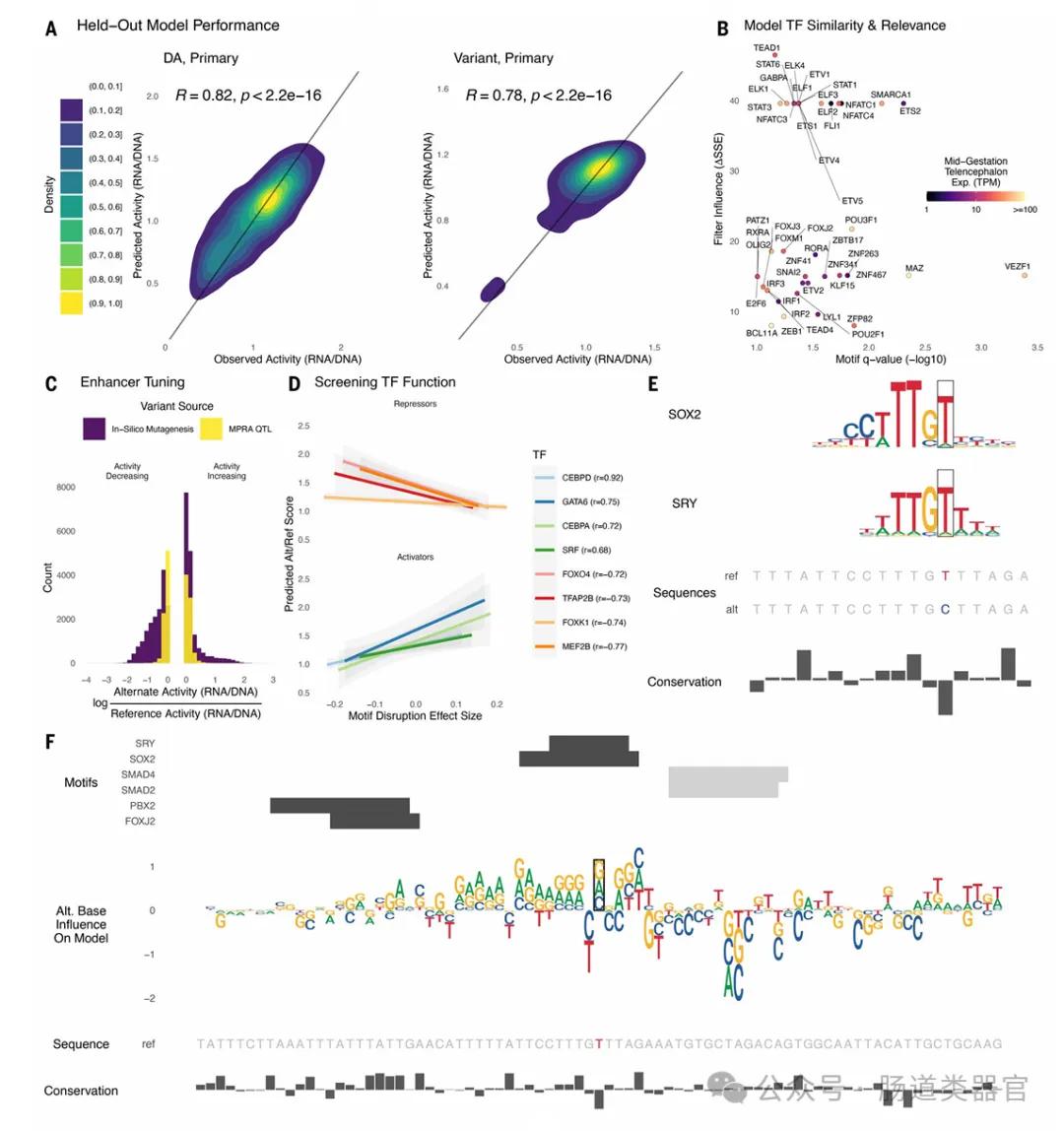
Deng et al. Research by combining lentivirus mass parallel report analysis technology and deep learning, gene regulation elements in human brain development, its value and significance is to understand the molecular mechanism of brain development provides a new perspective, reveals the gene regulation related to mental diseases, and provides a potential target for early diagnosis and treatment of mental illness. Furthermore, the study validates the feasibility of cerebral organoids as an in vitro model to simulate human brain development and demonstrates the potential of machine learning in predicting cell type-specific regulatory elements. By establishing a large-scale catalog of functional gene regulatory elements, this work not only advances our understanding of the regulatory code of human brain development, but also lays a solid foundation for future research on genomics, neurobiology, and mechanisms of related diseases.




